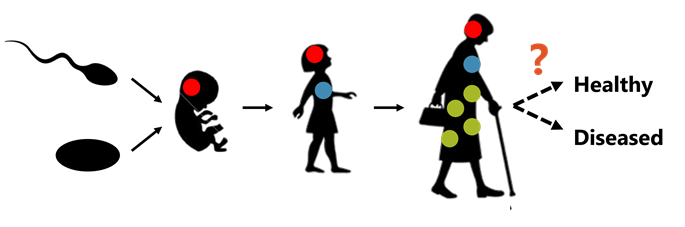
In the children’s game of telephone, one person whispers a message to a second person and then that person whispers the message to a third person and so on. At the end of the game, the original message is compared to last message. This game plays out on a colossal scale in your body over your entire life. The players are your cells, the message is your genome, and the message is passed from one cell to its daughter cells during cell replication. However, instead of a simple sentence or two, the message is as long as 2000 War and Peace novels! And, instead of playing the game with a small handful of friends, the game is played by every cell in your body (roughly 10,000,000,000,000,000 cells — the equivalent population of 1 million Earth populations!). It’s almost unbelievable that the scale of this game plays out in a person. Just like in the children’s game, the message starts to pick up errors over time: mutagenic processes like UV light and cigarette smoke distort the message. The mistakes can be trivial like changing the word from “small” to “little,” but they can also have grave consequences like changing the word from “love” to “hate.”
Currently, it’s unknown how many mistakes are made in the genome message over a person’s lifetime and what effect the mistakes have on their health.
To help answer these questions, we scanned the genome messages of ~1000 postmortem donors across many of their organs and looked for mistakes, or mutations. We learned that some tissues, like the esophagus and liver, acquire a lot of mutations whereas other tissues like brain, acquire fewer mutations. This made sense to us because esophagus and liver are exposed to many environmental toxins: here the cells must transmit the message in a noisy environment. A low number of mutations in the brain also makes sense because the brain is primarily composed of cells that don’t replicate: the cells only play at the beginning of the game when the genome message is relatively free from errors.
Since the telephone game is started immediately after conception, we were interested in knowing when the mutations occur during one’s lifetime. To our surprise, most of the mutations that occurred during gestation occurred at a very early and critical period of development called gastrulation. Furthermore, when we looked at what kind of mutations were made during gestation, we were again surprised to find that those mutations were predicted to be more harmful than the mutations that are associated with cancer and other diseases. The data showed patterns suggesting that the cells were actively preferring these potentially harmful mutations over neutral mutations — like a group of rogue telephone players purposely changing the meaning of the message. These results suggest that the mutations that occur over one’s lifespan have the potential to alter their health.
To ensure the health of the next generation, it’s critical that egg and sperm cells (also called germ cells) receive a pristine genome message. The body has several strategies for this, e.g., only letting these cells play at the very beginning of the telephone game when the genome message is less likely to have a corrupted message. We asked how accurate the game of telephone was for germ cells. Using mutations from several key points in the germ cell lifecycle, e.g., before puberty, after puberty, and birth of a child, we determined that the genome messages with potentially harmful mutations were likely removed over the course of the germ cell telephone game. We think this might be achieved through cell death, and spontaneous loss of pregnancy.
In the future, we hope this large database of mutations and mutation patterns will help scientists better understand and predict which corrupted genome messages are harmless and which turn into disease.
To learn more about this study, please check out our preprint:
Rockweiler NB et al. (2021). The origins and functional effects of postzygotic mutations throughout the human lifespan. BioRxiv. https://doi.org/10.1101/2021.12.20.473199